Biography
I am the founder of Efference and a PhD student in Computer Science at Princeton University, advised by Tom Griffiths. At Efference, I build cameras, specifically ones that replicate the robustness and efficiency of the human vision. At Princeton, I work on optimization and machine learning methods inspired by neuroscience, with recent interests in how physical and neurological development shape optimization landscapes—making human problems locally convex—and in neural architectures inspired by the fruit fly optic lobe.
Prior to Princeton, I did an undergraduate degree in Biochemistry, spent two years at Harvard Medical School studying visual decision-making (advised by Dr. Richard Born), and worked at NASA's Jet Propulsion Laboratory, developing new methods for converting CO2 to O2 on Mars (advised by Dr. Simon Jones). More: CV.
In my free time, I enjoy shooting, surfing, and sailing.
Selected Publications
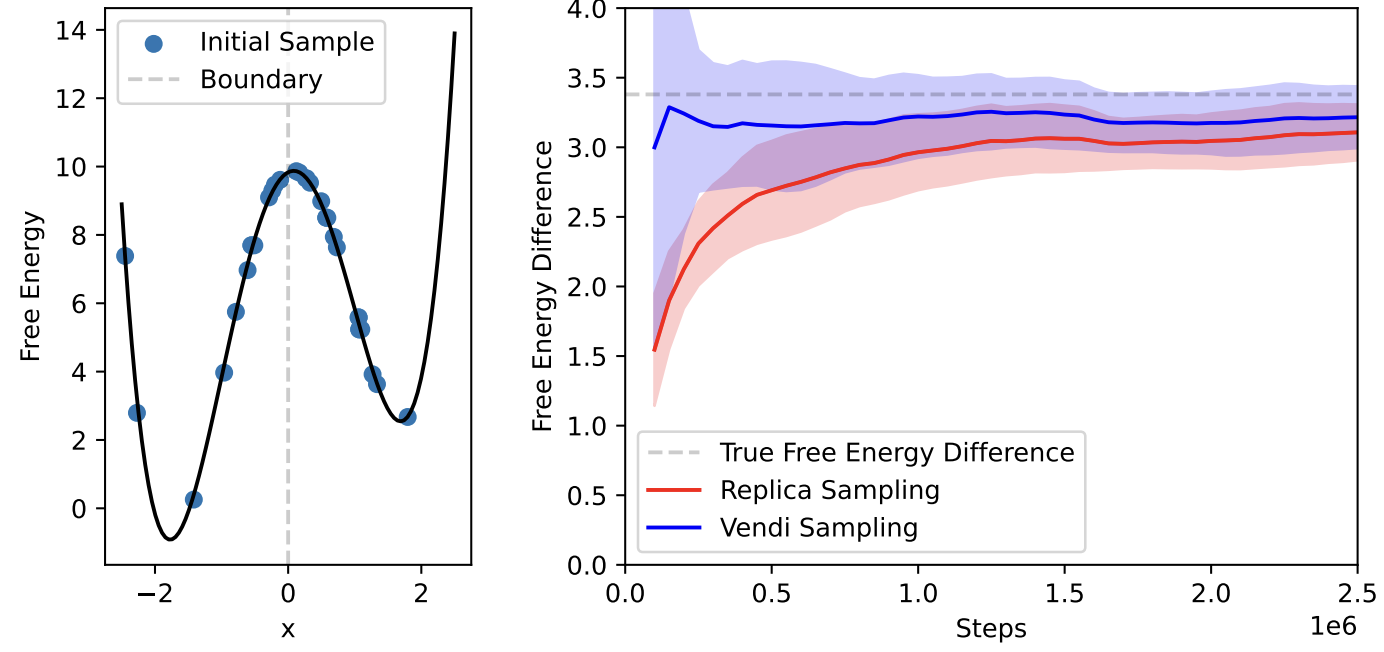
Journal of Chemical Physics.
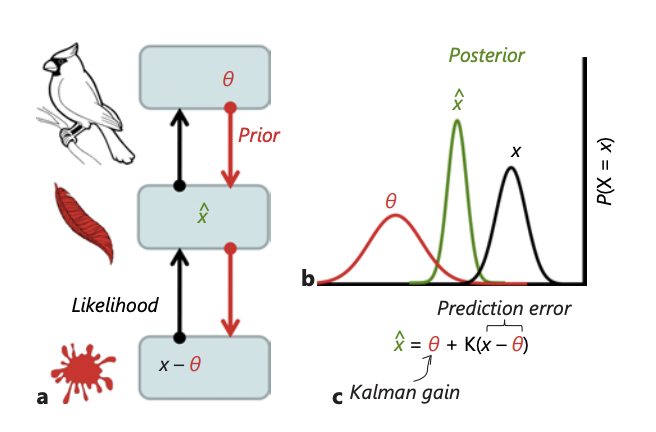
Illusions, Delusions, and Your Backwards Bayesian Brain: A Biased Visual Perspective.
Brain, Behavior and Evolution.